Jan 09, 2023
Author(s)
The field of biology is constantly advancing as researchers around the world work to uncover new insights into the mechanisms of life. With the vast amount of information being published on a daily basis, it can be a daunting task for biologists to stay up-to-date and extract relevant data for their research. This is where natural language processing (NLP) can play a crucial role. We are excited to introduce Sparse BioBERT, a state-of-the-art NLP model that is specifically fine-tuned on four unique biology-related datasets (described later).
By leveraging advanced machine learning techniques, BioBERT can quickly and accurately extract information from large volumes of unstructured text data. This includes information about specific medical conditions, treatments, and molecular structures. As you will see in the dataset descriptions, BioBERT can achieve this through various methods such as relation extraction, token classification (NER), or event extraction.
In this blog, we will discuss the capabilities of BioBERT and how it is revolutionizing the way biologists approach research. By enabling the rapid extraction of information from research literature, Sparse BioBERT will help biologists stay on the cutting edge of their field and, potentially, make new discoveries that were previously out of reach. We will also provide examples of how BioBERT can be successfully applied in real-world scenarios, and explore the potential for further advancements in the field of biology through the use of this powerful NLP tool.
Why BioBERT?
BioBERT is a pre-trained language model designed for NLP tasks in the biomedical domain. By training on a large corpus of biomedical literature, including articles from PubMed and PMC, BioBERT has a strong understanding of the language and concepts used in biomedicine. The BioBERT model has achieved state-of-the-art performance on a number of NLP tasks in the biomedicine domain, including named entity recognition, relation extraction, and question answering. It is widely used in the NLP community and has been applied to a variety of research projects and real-world applications in biomedicine, such as automated summarization of biomedical articles and analysis of electronic health records, and information extraction of entities in published research.
Currently, Neural Magic’s SparseZoo includes four biomedical datasets for token classification, relation extraction, and text classification. Before we see BioBERT in action, let’s review each dataset.
JNLPBA - Token Classification (NER)
The BioNLP / JNLPBA Shared Task 2004 involves the identification and classification of technical terms referring to concepts of interest in molecular biology such as DNA, RNA, protein, cell type, cell line. The JNLPBA corpus is distributed in IOB format, with each line containing a single token and its tag, separated by a tab character (similar to CoNLL). Sentences are separated by blank lines. Here are a few examples:
| Tokens | Labels |
| Number of glucocorticoid receptors in lymphocytes and their sensitivity to hormone action. | “O", "O", "B", "I", "O", "B", "O", "O", "O", "O", "O", "O", "O" |
| At the same time, total content of T lymphocytes was decreased 1.5-fold in peripheric blood. | “O", "O", "O", "O", "O", "O", "O", "O", "B", "I", "O", "O", "O", "O", "O", "O", "O" |
You can view the model cards for the sparse and dense baseline models fine-tuned on JNLPBA.
BC2GM - Token Classification (NER)
The BioCreative II Gene Mention Recognition (BC2GM) dataset contains data in which participants are asked to identify a gene mentioned in a sentence by giving its start and end characters. The training set consists of a set of sentences and, for each sentence, a set of gene mentions (GENE annotations). Here are a few examples:
| Tokens | Labels |
| Physical mapping 220 kb centromeric of the human MHC and DNA sequence analysis of the 43-kb segment, including the RING1, HKE6,and HKE4 genes. | "O", "O", "O", "O", "O", "O", "O", "B", "I", "O", "O", "O", "O", "O", "O", "O", "O", "O", "O", "O", "O", "B", "O", "B", "O", "O", "B", "I", "O" |
| Id-1H and Id-2H seem to be human homologues of mouse Id-1, and Id-2, respectively, and have potential to encode 154 and 135 amino acid proteins. | "B", "I", "I", "O", "B", "I", "I", "O", "O", "O", "O", "O", "O", "B", "I", "I", "I", "O", "B", "I", "I", "O", "O", "O", "O", "O", "O", "O", "O", "O", "O", "O", "O", "O", "O", "O" |
You can view the model cards for the sparse and dense baseline models fine-tuned on BC2GM.
ChemProt - Relation Extraction
ChemProt is a disease chemical biology database, which is based on a compilation of multiple chemical–protein annotation resources as well as disease-associated protein–protein interactions (PPIs). Here are a few examples:
| Sequences | Labels |
| @CHEMICAL$ antagonistic effect of estramustine phosphate (EMP) metabolites on wild-type and mutated @GENE$. | CPR:6 |
| We investigated, by displacement of @CHEMICAL$ in the presence of 2.5 mM of triamcinolone acetonide, the binding of estramustine phosphate and its metabolites, estramustine, estromustine, estrone, and beta-estradiol as well as other antiandrogen agents (including alpha-estradiol, bicalutamide, and hydroxyflutamide) to the mutated @GENE$ (m-AR) in LNCaP cells and to the wild-type androgen receptor in wild-type AR cDNA expression plasmid (w-pAR0) cDNA-transfected HeLa cells. | False |
You can view the model cards for the sparse and dense baseline models fine-tuned on ChemProt.
DDI - Text Classification
The DDI Extraction 2013 task concerns the recognition of drugs and extraction of drug–drug interactions that appear in biomedical literature abstracts. Here are a few examples:
| Sequences | Labels |
| @DRUG$: Absorption of a single dose of Myfortic was decreased when administered to 12 stable renal transplant patients also taking @DRUG$-aluminum containing antacids (30 mL): the mean Cmax and AUC(0-t) values for MPA were 25% and 37% lower, respectively, than when Myfortic was administered alone under fasting conditions. | DDI-false |
| Antacids: Absorption of a single dose of @DRUG$ was decreased when administered to 12 stable renal transplant patients also taking @DRUG$-aluminum containing antacids (30 mL): the mean Cmax and AUC(0-t) values for MPA were 25% and 37% lower, respectively, than when Myfortic was administered alone under fasting conditions. " | DDI-mechanism |
You can view the model cards for the sparse and dense baseline models fine-tuned on DDI.
Unlock the Knowledge of ArXiv Abstracts With Sparse BioBERT
Using biomedical named entity recognition (BioNER) as part of the researcher’s text mining initiatives can help researchers stay abreast of emergent relationships of entities in biomedicine. As such, BioNER is often conducted as the first step toward relation detection (which identifies how two entities relate to each other). This downstream task offers researchers the ability to identify novel relationships in the latest research on a specific topic of interest. However, finding novel relationships at the rate of the recent spike in published research can be very time-consuming if the researcher uses a dense state-of-the-art Transformer model.
To demonstrate the performance of Sparse BioBERT in the molecular biology domain, we will conduct inference on a subset of the large ArXiv dataset focused on papers tagged with the "Quantitative Methods (q-bio.QM)" tag. This tag identifies biomedical literature showing experimental, numerical, statistical, and mathematical contributions of value to biology.
The dataset consists of more than 150,000 tokens, and we will perform named entity recognition on each abstract. To do this, we will use a sparse BioBERT model that has been fine-tuned on the JNLPBA dataset for the token classification task.
Let’s first install the DeepSparse and datasets libraries:
pip install deepsparse datasetsRun the following Python script to download the pruned and quantized BioBERT, our subset of the ArXiv dataset, and pass in the abstracts for token classification:
TIP: To run inference on the dense variant, substitute the following zoo stub to the `model_path`: zoo:nlp/token_classification/biobert-base_cased/pytorch/huggingface/jnlpba/base-none
from deepsparse import Pipeline
from datasets import load_dataset
from time import perf_counter
import pprint as pp
dataset = load_dataset("zeroshot/arxiv-biology")
abstracts = dataset["train"]["abstract"]
model_path = "zoo:nlp/token_classification/biobert-base_cased/pytorch/huggingface/jnlpba/pruned90_quant-none"
pipeline = Pipeline.create(
task="token-classification",
model_path=model_path
)
start = perf_counter()
inference = pipeline(abstracts)
pp.pprint(inference.predictions)
end = perf_counter()
delta = end - start
print("secs: ", delta)The following is a printout of the DeepSparse pipeline showing the ‘B’ and ‘I’ tagged tokens of a single abstract:
[[TokenClassificationResult(entity='B', score=0.9676008820533752, index=50, word='m', start=232, end=233, is_grouped=False),
TokenClassificationResult(entity='I', score=0.9225506782531738, index=51, word='##RNA', start=233, end=236, is_grouped=False),
TokenClassificationResult(entity='I', score=0.8929973244667053, index=52, word='##s', start=236, end=237, is_grouped=False),
TokenClassificationResult(entity='B', score=0.6662591695785522, index=56, word='gene', start=259, end=263, is_grouped=False),
TokenClassificationResult(entity='I', score=0.7430179715156555, index=57, word='products', start=264, end=272, is_grouped=False),
TokenClassificationResult(entity='B', score=0.9038915038108826, index=92, word='m', start=434, end=435, is_grouped=False),
TokenClassificationResult(entity='I', score=0.8745383024215698, index=93, word='##RNA', start=435, end=438, is_grouped=False),
TokenClassificationResult(entity='B', score=0.92357337474823, index=97, word='protein', start=460, end=467, is_grouped=False),
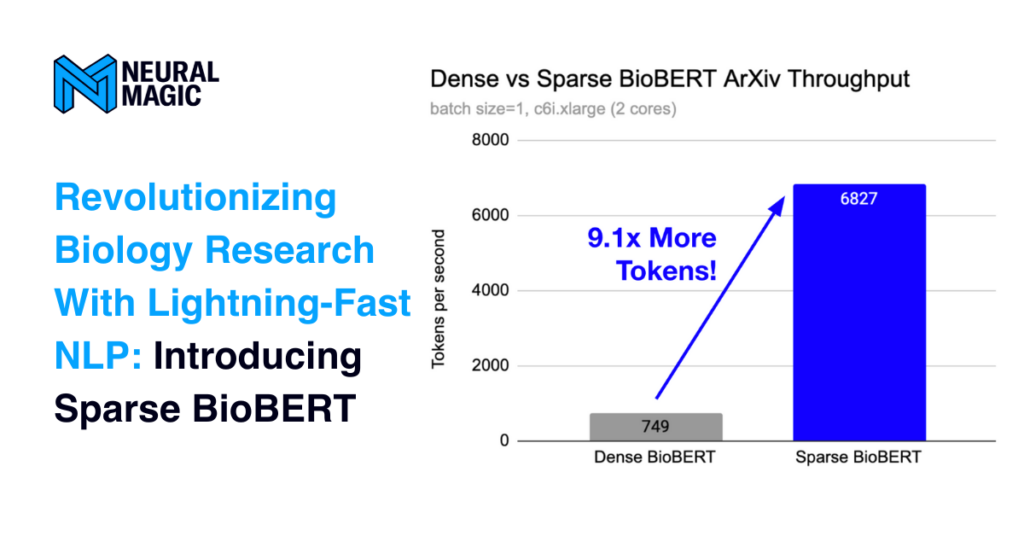
TokenClassificationResult(entity='I', score=0.8777768611907959, index=98, word='molecules', start=468, end=477, is_grouped=False)]]Additionally, we benchmarked the sparse model against the dense variant using DeepSparse on an AWS c6i.xlarge instance with two cores to show the difference in the sparse and quantized model.
total abstracts: 1,277 | total tokens: 157,154 | batch=1
| Model | Engine | Instance | Latency (secs) | Tokens per second |
| Dense BioBERT | DeepSparse | c6i.xlarge (2 cores) | 209.71 | 749.387 |
| Sparse BioBERT | DeepSparse | c6i.xlarge (2 cores) | 23.02 | 6826.846 |
Final Thoughts
We introduced Neural Magic’s new sparse BioBERT fine-tuned on four unique biology-related datasets. In particular, the JNLPBA dataset was benchmarked against its dense variant on the open-sourced ArXiv-biology dataset extracted from the larger ArXiv dataset. The performance improvement when computing more than 100K tokens was over 9 times faster than the dense variant.
For more on Neural Magic’s open-source codebase, view the GitHub repositories (DeepSparse and SparseML). For Neural Magic Support, sign up or log in to get help with your questions in our community Slack. Bugs, feature requests, or additional questions can also be posted to our GitHub Issue Queue.