Nov 16, 2023
Author(s)
The AI space is abuzz with large language models (LLMs), but using them locally is a challenge due to their enormous size. Organizations that want to use these models for applications such as question answering must either invest in expensive cloud infrastructure or use closed-source models. By using closed-source models, companies also give up their privacy and pay for APIs. But, what if there is another option?
An alternative is to make open-source LLMs small enough to run on a laptop without negatively impacting the model’s accuracy. Neural Magic provides the tools to optimize LLMs for CPU inference, which means you can run LLMs on your laptop without a GPU. Fast inference on CPUs is possible with DeepSparse—Neural Magic's inference runtime that takes advantage of sparsity to accelerate neural network inference. We also provide various optimized open-source LLMs on our SparseZoo that users can download and use immediately.
In this blog post, you will learn how to use DeepSparse and open-source LLMs, such as MPT and Llama, to build a medical chatbot that runs on your laptop without a GPU. The chatbot sources answers to questions from the Gale Encyclopedia of Medicine. This LangChain-based chatbot also cites the pages from where the answers are retrieved from. Considering the amount of information in this medical encyclopedia, this chatbot is an efficient way to learn from the textbook. The medical chatbot can also suggest possible illnesses based on a set of symptoms. Check out the bot in action below:
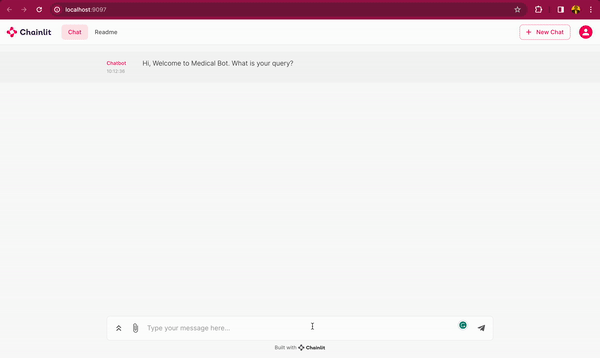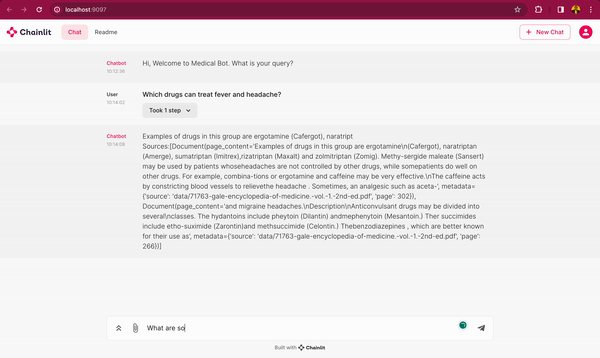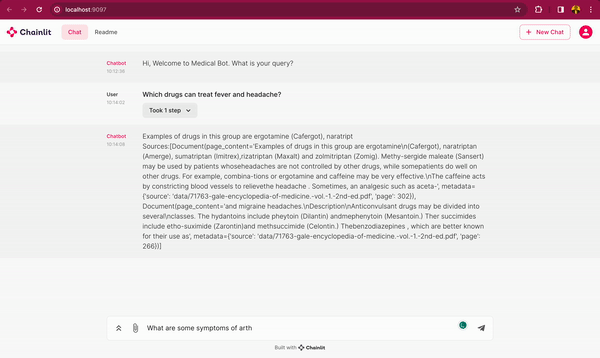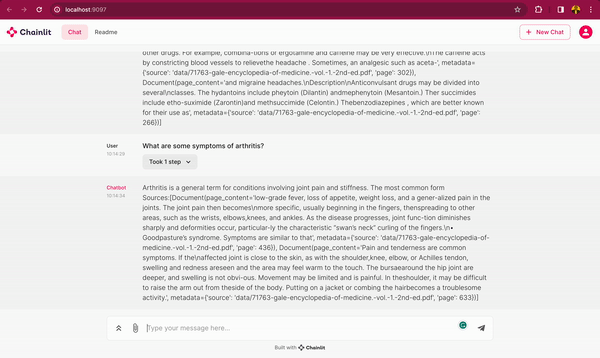
You can also follow along with the full code in our examples repository.
Data Ingestion
Large language models have limited context length, which means that you can’t pass the entire medical encyclopedia to the LLM. The solution is to:
- Load the PDF.
- Split the text into multiple chunks.
- Create word embeddings for each chunk.
- Store the embeddings in a vector database such as Faiss.
Here’s how to do that in code:
from langchain_community.embeddings import HuggingFaceEmbeddings
from langchain_community.vectorstores import FAISS
from langchain_community.document_loaders import PyPDFLoader, DirectoryLoader
from langchain.text_splitter import RecursiveCharacterTextSplitter
DATA_PATH = 'data/'
DB_FAISS_PATH = 'vectorstore/db_faiss'
# Create vector database
def create_vector_db():
loader = DirectoryLoader(DATA_PATH,
glob='*.pdf',
loader_cls=PyPDFLoader)
documents = loader.load()
text_splitter = RecursiveCharacterTextSplitter(chunk_size=500,
chunk_overlap=50)
texts = text_splitter.split_documents(documents)
embeddings = HuggingFaceEmbeddings(model_name='sentence-transformers/all-MiniLM-L6-v2',
model_kwargs={'device': 'cpu'})
db = FAISS.from_documents(texts, embeddings)
db.save_local(DB_FAISS_PATH)
if __name__ == "__main__":
create_vector_db()
Load the LLM on Your CPU
With the vector database in place, the next step is to set up the LLM locally. DeepSparse provides a LangChain interface with configurable parameters such as:
- The path to the model which can be a Hugging Face path or SparseZoo stub.
- The maximum number of tokens you’d like the model to generate—the bigger the number, the longer it takes to generate the final output.
from langchain_community.llms import DeepSparse
import os
MODEL_PATH = os.environ.get("MODEL_PATH") or "hf:neuralmagic/mpt-7b-chat-pruned50-quant"
def load_llm():
# Load the locally downloaded model here
llm = DeepSparse(
model=MODEL_PATH,
model_config={"sequence_length": 2048, "trust_remote_code": True},
generation_config={"max_new_tokens": 300},
)
return llm
llm = load_llm()
Passing the model path via an environment variable will allow you to initialize the application with different models at runtime.
Create a Prompt Template
Next, create a prompt that will dictate the chatbot's behavior. For example, you can explicitly direct the model to not make up an answer if it can’t be found in the given text––this is important in preventing hallucination.
from langchain import PromptTemplate
custom_prompt_template = """Use the following pieces of information to answer the user's question.
If you don't know the answer, just say that you don't know, don't try to make up an answer.
Context: {context}
Question: {question}
Only return the helpful answer below and nothing else.
Helpful answer:
"""
def set_custom_prompt():
# Prompt template for QA retrieval for each vectorstore
prompt = PromptTemplate(
template=custom_prompt_template, input_variables=["context", "question"]
)
return prompt
Set Up a Retrieval QA Chain
LangChain provides the RetrievalQA to obtain answers given a query. The chain parameters include:
- The LLM model, in this case, the DeepSparse model.
- The type of chain, using
stuff, gathers all the documents and makes one call to the LLM. - The retriever that should be used for fetching documents and passing them to the LLM.
- Whether to return the source documents.
- Extra keyword arguments, for example, the prompt.
# Retrieval QA Chain
def retrieval_qa_chain(llm, prompt, db):
qa_chain = RetrievalQA.from_chain_type(
llm=llm,
chain_type="stuff",
retriever=db.as_retriever(search_kwargs={"k": 2}),
return_source_documents=True,
chain_type_kwargs={"prompt": prompt},
)
return qa_chain
Question Answering With Faiss
Next, we need to obtain answers from the chatbot after submitting a question. The process for getting the answers works as follows:
- Input a question on the chatbot interface.
- Create an embedding for the question.
- Compare the embedding of the question with the embeddings in the vector database.
- Return the top K embeddings that are similar to the question’s embeddings.
- Pass these embeddings to the LLM for question-answering.
- Return response from LLM.
Here’s how to implement this in code:
# QA Model Function
def qa_bot():
embeddings = HuggingFaceEmbeddings(
model_name="sentence-transformers/all-MiniLM-L6-v2",
model_kwargs={"device": "cpu"},
)
db = FAISS.load_local(DB_FAISS_PATH, embeddings)
qa_prompt = set_custom_prompt()
qa = retrieval_qa_chain(llm, qa_prompt, db)
return qa
# Output function
def final_result(query):
qa_result = qa_bot()
response = qa_result({"query": query})
return response
Set Up a Chainlit Application
With all the building blocks in place, you can use Chainlit to build a simple interface for the LLM application. The application does the following:
- Prints a welcome message.
- Initializes the retrieval chain on start.
- Allows you to type a question.
- Returns the answer and the corresponding sources.
# Chainlit code
@cl.on_chat_start
async def start():
chain = qa_bot()
msg = cl.Message(content="Starting the bot...")
await msg.send()
msg.content = "Hi, Welcome to Medical Bot. What is your query?"
await msg.update()
cl.user_session.set("chain", chain)
@cl.on_message
async def main(message: cl.Message):
chain = cl.user_session.get("chain")
cb = cl.AsyncLangchainCallbackHandler(
stream_final_answer=True, answer_prefix_tokens=["FINAL", "ANSWER"]
)
cb.answer_reached = True
res = await chain.acall(message.content, callbacks=[cb])
answer = res["result"]
sources = res["source_documents"]
if sources:
answer += f"\nSources:" + str(sources)
else:
answer += "\nNo sources found"
await cl.Message(content=answer).send()
Start the application with your preferred LLM:
MODEL_PATH="hf:neuralmagic/mpt-7b-chat-pruned50-quant" chainlit run model.py -w
The application will be available at http://localhost:8000.
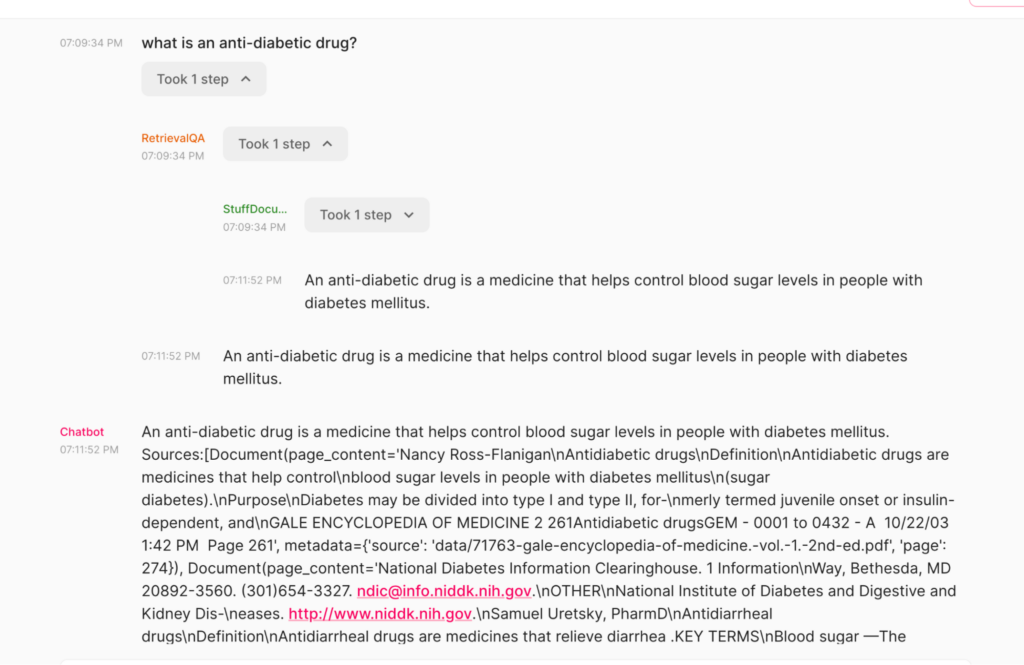
Final Thoughts
This blog proves it is possible to build LLM applications locally using DeepSparse which offers accelerated inference on CPUs. You can run the latest large language models on your computer without having to configure special hardware when you use optimized open-source LLMs. Whether you are building hobby projects to explore state-of-the-art LLMs or deploying them in production environments where speed and cost are critical, DeepSparse has a solution for you.
In addition to DeepSparse, for accelerated inference on CPUs, Neural Magic provides other tools and resources to make your LLM development a success. Check out our: